
CISC-471 Computational Biology
Fall 2012
 |
CISC-471 Computational Biology |
 |
Last updated: October 31, 2012
Announcements
• Make sure to submit your project proposals BEFORE Monday, November 5, 2012.
• The midterm exam will be held in Nicol Hall, Room 321 on Monday, October 22 from 8:30-10:30.
• Due to popular demand, there is a ~3-hour extension for assignment 2. It is now due on Friday, Oct 12 at 11:59pm, instead of 9:00pm. Any submissions uploaded on or after 12:01am will be considered late.
Instructor |
Hazem R. Ahmed |
|
|
Office |
Goodwin 621 |
Office Hours |
By Appointment (or after class) |
Class Time |
Tuesdays 08:30-09:30 Wednesdays 10:30-11:30 Fridays 09:30-10:30 |
Class Room |
Botterell Hall, Room B148. |
Teaching Assistant |
Thomas Vaughan |
Tutorial/Lab Time |
Mondays 08:30-10:30 |
Tutorial/Lab Room |
Goodwin Hall, Room 248. |
Course Description
Advanced computational approaches to the problems in molecular biology. Techniques and algorithms for pair-wise and multiple sequence analysis and alignment; molecular databases; phylogenetic trees construction, protein structure prediction and alignment; microarray technology, gene expression data analysis and interpretation.
This is a required course for Biomedical Computing students. The official prerequisites for CISC 471 are BCHM 315 (or BIOL 334); CISC 271, 352; MBIO 218 (or 318).
Course Texts
•Recommended Textbooks:
The course will not strictly follow a particular textbook, so students are encouraged to attend the lectures. However, course materials will rely mostly on the following textbooks.
•Reference Textbooks:
The following reference books are just for additional (optional) background reading on relevant topics discussed in the class.
Course Outline
Course materials and assignments are managed using Queen’s Moodle
Guest Lecturers
• Dr. Donald Forsdyke, Professor in Biochemistry, Department of Biomedical and Molecular Sciences (Chargaff's Rules and Evolutionary Bioinformatics - Week 2)
• Dr. Paul Young, Professor in Biology, Department of Biology (Substitution Matrices - Week 3)
• Dr. Brent Wathan, Post-Doctoral Fellow in Biochemistry, Department of Biomedical and Molecular Sciences (Protein Folding - Week 8)
• Dr. Harriet Feilotter, Associate Professor in Pathology, Department of Pathology and Molecular Medicine (DNA Microarray Technology and Next-Generation Sequencing - Week 10)
• Dr. Paulo Nuin, Assistant Professor in Pathology, Department of Pathology and Molecular Medicine (Gene Expression Data Interpretation - An Application of Bioinformatics to Cancer Research - Week 11)
Assessment and Grading Scheme
• Midterm Exam – 25% (Monday, October 22, 8:30-10:30)
Assignment Submission
Web and programming exercises are to be uploaded in Moodle by 9pm EST on the due date specified, and problem solving assignments are to be handed in class on the due date specified.
Late Policy
Each assignment is due on the due date specified and will be penalized 10% for each 24h late, up to a maximum of 3 days (including weekend-days), after which it cannot be accepted.
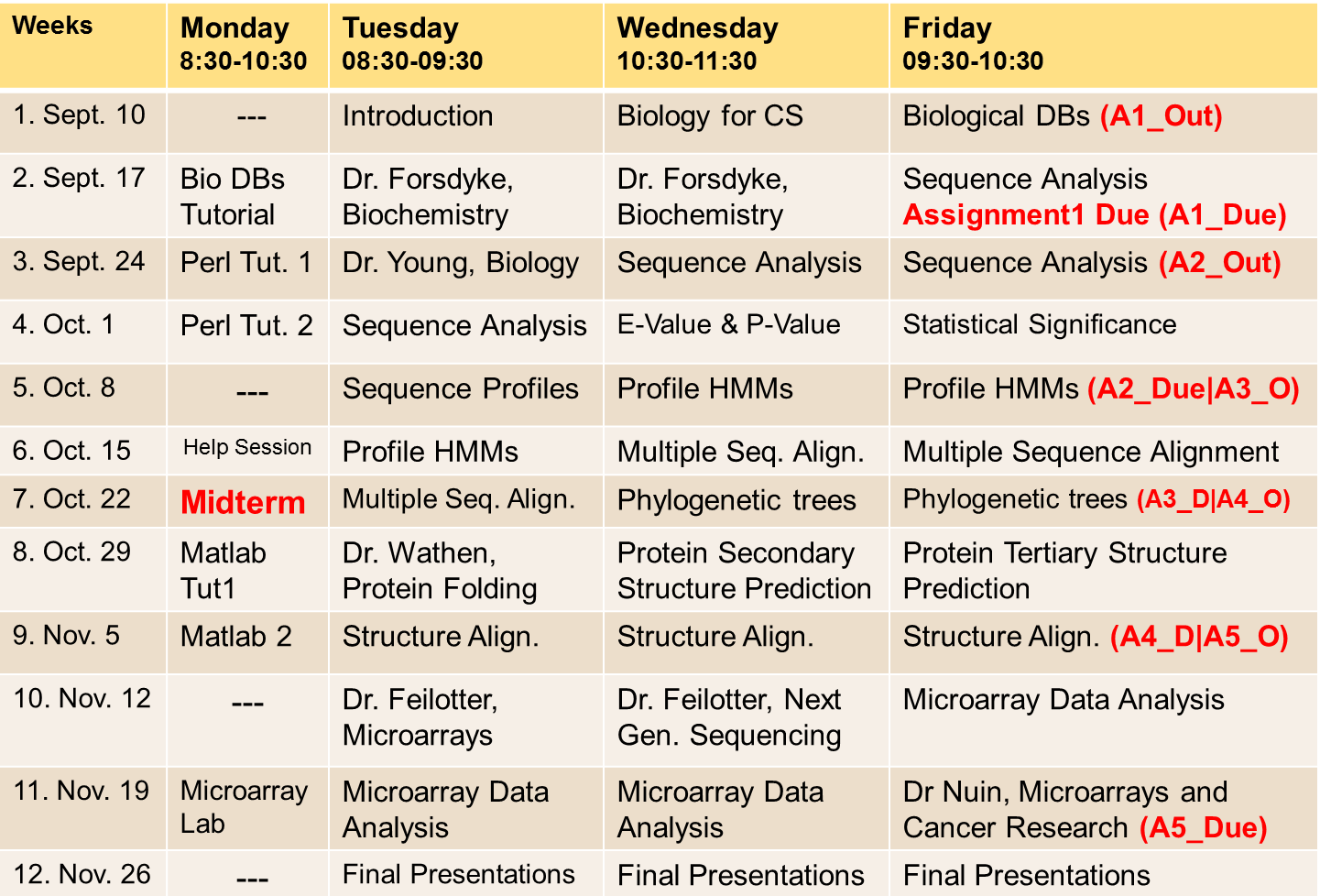
Course Schedule
The details of the course weekly schedule are for orientation purposes only. This does not apply to the Midterm date or the dates of the Final Presentations, which should not change without prior notice.
|
Academic Integrity
Academic integrity is constituted by the five core fundamental values of honesty, trust, fairness, respect and responsibility. These values are central to the building, nurturing and sustaining of an academic community in which all members of the community will thrive. Adherence to the values expressed through academic integrity forms a foundation for the "freedom of inquiry and exchange of ideas" essential to the intellectual life of the University (see the Senate Report on Principles and Priorities.) Students are responsible for familiarizing themselves with the regulations concerning academic integrity and for ensuring that their assignments conform to the principles of academic integrity.
Students are responsible for familiarizing themselves with the regulations concerning academic integrity and for ensuring that their submitted work conforms to the principles of academic integrity. Information on academic integrity is available in the Arts and Science Calendar (see Academic Regulation 1), on the Arts and Science website and from the instructor of this course.
Departures from academic integrity include plagiarism, use of unauthorized materials, facilitation, forgery and falsification, and are antithetical to the development of an academic community at Queen's. Given the seriousness of these matters, actions which contravene the regulation on academic integrity carry sanctions that can range from a warning or the loss of grades on an assignment to the failure of a course to a requirement to withdraw from the university.
While there will be no tolerance for violation of Regulation 1, student "conceptual" discussions and brainstorming about course assignments and topics are encouraged.
Accommodation Requests
Please let your instructor know if you need any special accommodations.